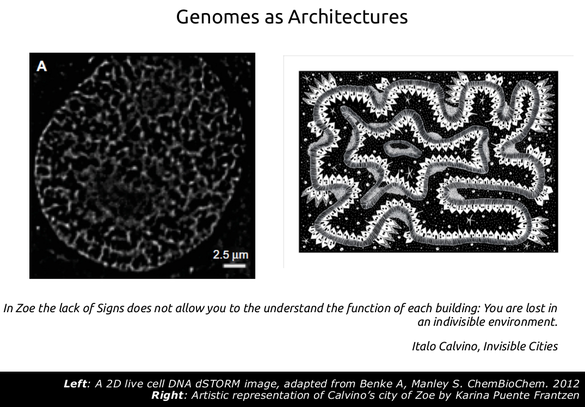
In our group, one of our most active interest is the evolution of genome architecture in an almost literal sense of the term. Thus we try to envision the genomes, especially those of eukaryotes, as structures with an inherent organization, which we cannot (obviously) yet fully decipher. In this respect, we have been studying genomic characteristics for which we can get a lot of data, such as gene expression, regulation, chromatin structure, conservation etc under the lens of their spatial distribution in the genome in both linear and three-dimensional space.
So earlier this year, starting from an already published dataset, on which we had worked before, we tried to answer a simple question: Do genes with similar properties cluster in the linear space of eukaryotic genomes? This is obviously not a new question. In fact since the very first gene expression analysis papers, people had already realized that gene expression is correlated with gene position (see for example here and here). Soon after came analyses that showed that the proximity between genes was also associated with common regulation (like e.g. in this great paper). In our work we tried to combine all of the above studied properties, plus some more, to show that genes in a simple eukaryotic genome like yeast tend to cluster in particular domains with clear structural and organizational characteristics. Your can read more in our paper here and two relevant blog posts here and there, but the main point is that the yeast genome looks to have involved in such a way, so that their is a "division of labour" of genes in distinct neigbourhoods of the genome, the clearest of which is a distinction between the edges and the centers (i.e. the centromeres) of the chromosomes.
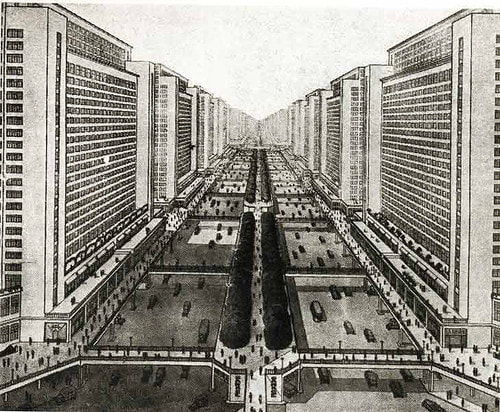
Back in the 20s and 30s these concepts were highly popular as can be seen in Le Corbusier's Ville Radieuse which was the inspiration for Oscar Niemeyer planning of the city of Brazilia. Utopian architecture has also inspired Italo Calvino, who in one of his Invisible Cities describes the city of Zoe as a place where "the lack of signs does not allow you to understand the function of each building; your are lost in an indivisible environment". The Ville Radieuse (shown below) or an artistic representation of Zoe (above right) are examples of how the lack of landmarks may appear at the same time all-encompassing but also hard to navigate. In fact, utopian architectures are nowadays only used in particular cases such as airport terminals or metro stations, which you can only navigate when assisted by additional signals (gate numbers, names of stations) and very often the constant help of the voice of an announcer. Most modern city landscapes are not like this. They have particular landmarks: parks, boulevards, avenues and buildings that are distinct from each other, which gives them a symbolic character. They are not designed to be thus but have probably "evolved" (in a relaxed sense of the term) like this through the gradual aggregation of elements. They are in this sense easier to "parse" by being able to distinguish the City Hall from the General Hospital, the University campus from an industrial zone etc.
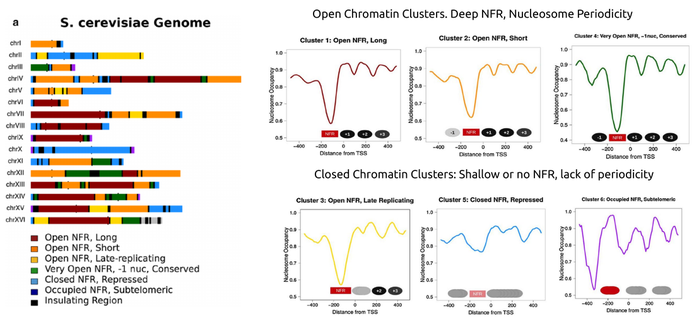
By obtaining publicly available nucleosome occupancy data we looked into the patterns of nucleosome positioning around the transcription start sites of the genes contained in each of the defined domains. Even though there is a general pattern of nucleosome positioning in yeast genes, with a clear nucleosome free-region (NFR) located exactly upstream of the TSS, we were surprised to see that different topological domains harboured genes with very different nucleosomal architectures. We were, moreover, able to cluster yeast TADs in six different categories, according to their patterns of nucleosome positioning. In the Figure below, one can see how radically different the short-scale chromatin structures can be. A clear distinction between a more "open" and a more "closed" chromatin conformation can be directly linked to regulatory and functional properties of these domains. The main point though, was that we were able to answer our initial question: There appear to be structurally-defined landmarks that distinguish different "neighbourhoods" in the yeast genome. Even though, a large number of additional aspects can be thought of (enrichment in transcription factors, epigenetic marks etc), nucleosome positioning patterns represent a more general characteristic that is directly linked (and possibly affecting) most others.
 RSS Feed
RSS Feed
