But since TADs had not (at the time) been reported in yeast, one of the main criticisms that we received during the review process was directed at this analysis. The reviewer's comment was:
"Authors declare the existence of TAD-like globules in budding yeast. However, these kinds of structures have, so far, never been detected in Saccharomyces cerevisiae. If the authors want to establish the existence of such TAD like structures, they must reinforce their analysis."
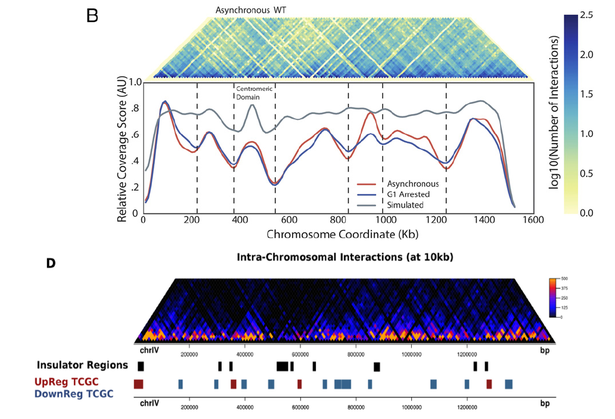
At the time, we were eager to get the paper accepted and so we down-played the original term "TAD-like" in "insulated domains" in the final version. Figure 4D remained though and was further supported by a number of analyses that showed the robustness of the boundaries upon different normalization strategies and the lack of LTRs in these regions. In our view, it mattered little to get the message of "TADs also exist in yeast" across, as our main interest was to show the tendency for spatial clustering of genes.
What is more important, the properties that are shared by the TAD boundaries in Eser et al. are matching our observations in many respects, as they are shown to be enriched in transcription activity (as originally shown by the group of Sergei Razin) and activating histone marks. Above all, Eser et al. report that regions between the defined TADs are significantly overlapping areas of topoII depletion, which constitutes an immediate link to our finding that genes that tend to be up-regulated by topoII inactivation are enriched in insulated regions (remember this was our way to call TAD boundaries, without using the term "TAD"). Thus, even though one of the main arguments in Eser et al. is that TADs in yeast are mostly related to DNA replication than transcription, it seems that you cannot really do away with transcriptional effects in relation to chromatin organization, especially in a unicellular eukaryote genome where the two processes are expected to be more tightly connected.
In closing, we can now be confident that TADs, or TAD-like domains if you will, do exist in yeast and that they are inherently related to both DNA replication and transcription (even though perhaps indirectly). Our observations under topological stress lie in the interphase between the two processes as torsional stress accumulation inevitably affects the DNA replication process. Even more interesting, in our view, is the fact that the embedded constraints in the organization of genome architecture are reflected on the evolution of gene distribution along chromosomes, but then again we have already discussed this elsewhere. One last point that we can make is that it is reassuring to see you can constructively argue with a reviewer (provided the reviewer's sanity) if your hypothesis is solid and supported by the data and that it is always nice to see you were right in the first place, even though sometimes courtesy towards a reviewer obliges you to be less audacious in the choice of terminology.
 RSS Feed
RSS Feed
