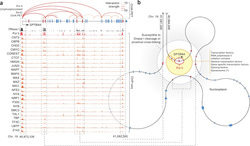
The main idea: Exons have been recently shown to be spliced co-transcriptionally, which means that splicing takes place while the DNA is still being transcribed. The implications of the coupling of these process are various. A very important one is the definition of exons to be spliced-in (thus retained in the initial mRNA transcript) taking place through the interaction of DNA regions in the three dimensional space. The authors examine this possibility through the combination of chromosome interactions in the three dimensional space, DNAseI hypersensitivity assays and RNA sequencing.
The data used: The authors performed a DNAaseI digital footprinting assay to obtain information of chromatin accessibility throughout the genome, ChIA-PET in order to determine distant interactions at chromatin level coupled with RNA sequencing in order to obtain information on the structure of transcripts (effectively which exons are included in mRNAs)
The analysis: After going through all necessary steps for the initial mapping and normalization of sequencing data, the analyses was quite straight-forward. The authors looked for exons that appear to be alternatively spliced in specific tissues (that is non-constitutive exons) to demonstrate that they are positioned in the proximity of promoters and are thus affected by DNAseI. Thus they hypothesize that promoter and enhancer activity can be exerted at the level of splicing by effectively modulating genome proximity. In other words, promoters and enhancers are physically interacting with those exons that will be alternatively spliced in mRNA transcripts and mark them accordingly.
What's next: One of the most interesting aspects of this work is that it extends the notion of "transcription factories" according to which genes that are to be transcribed are organized to concrete entities by approaching each other in physical space. According to Mercer et al., findings this view may be further extended to the level of gene, where exons to be alternatively spliced are co-localized with promoter and enhancer elements and thus gain the necessary "markings" that will eventually define their inclusion or not. Further experimental validation will prove this exciting new feature of gene expression regulation at the level of mRNA transcripts.
Read more: On the DNAaseI digital footprinting method (Neph et al., 2012) and on chromosome conformation capture approaches (Bau et al., 2010) and ChIA-PET (Fullwood et al, 2009). Read more on the transcriptional-factory hypothesis in (Cook, 2010)
 RSS Feed
RSS Feed
