In a recent paper in Genome Research, Tess Jeffers and Jason Lieb, bring back the notion of fragile nucleosomes, this time through conventional "good-old" MNase-Seq. In a nutshell, what Jeffers and Lieb did is not very different from early works on the concept of fragile nucleosomes (i.e. nucleosomes that can be digested by MNase and are thus "lost" from MNase maps) in the sense that they use differential timing MNase digestion, separating the output and treating a specific set of sequences sizes that comes from low digestion times as the "fragile" fraction.
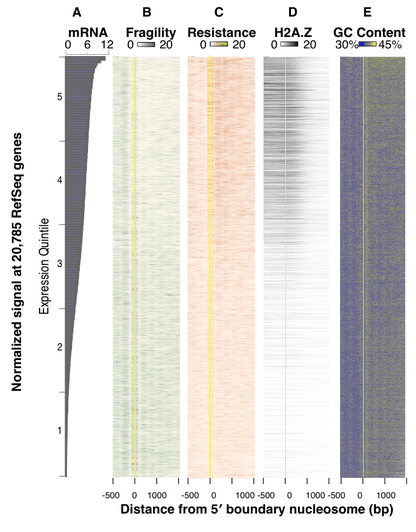
Through a rather straightforward analysis of differential nucleosome positioning in C. elegans embryos, the authors were able to recap most of what we knew about nucleosomes already. Fragile nucleosomes are AT-rich, enriched in promoters of low-expression genes, lacking enrichments in regulatory activating marks and isoforms such as H2A.Z. This reassuring(?) image is summarized in the Figure below (Figure 4a in their paper), where fragility and resistance correspond to the fragile and "well-positioned" fractions of the bulk nucleosomes.
Still, as this little story develops it would be interesting to see if the chemical-mapping method was just a firework or is here to make a difference.
 RSS Feed
RSS Feed
