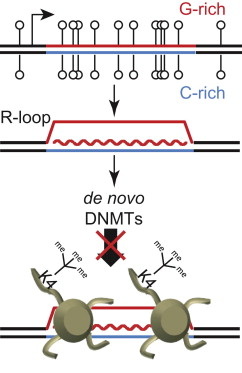
In this work Ginno et al., elaborate more on a previously published work, where they showed that GpG islands coincide with genomic regions that share an intrinsic pattern of "base overloading", showing increased representation of either Gs or Cs for long tracts of DNA. These patterns, known as GC (or more generally "nucleotide") skews, are representative -the authors show- not only of CpG islands but also of the formation of R-loops, molecular hybrid structures, in which the newly synthesized RNA re-anneals to the DNA to form a sort of triplex. In this very interesting piece of innovative research, the authors further show a link between the location and pattern of the skews and the properties of CpG islands
The data used: Human ES cells were profiled for R-loop formation with DRIP-Seq, a novel technique established by the same group in the aforementioned previous work. GC skewed regions were defined with the application of a custom R-script called SkewR. DNA methylation and gene expression data were obtained from previous published works
The analysis: The authors clustered all promoters of the human genome based on the existence of a) strong b) weak c) no or d) inverse GC skews (inverse meaning Cs outnumbered Gs instead of the opposite) and then went on to characterize specific properties of both the promoters and the downstream genes. Not surprisingly, they found that a strong (normal or inverse) GC-skew pattern coincided with increased expression but were also able to demonstrate that DNA methylation occurs much less frequently in promoters with GC skew compared to those with an even nucleotide composition. They were also able to correlate GC skews with the formation of R-loops genome-wide. Their results suggest a molecular mechanism (R-loop formation) that is guided by the underlying nucleotide composition (DNA skews) and results in an epigenetic effect (resistance to DNA methylation).
What's next: This work highlights the -often forgotten- role of underlying DNA sequence composition in molecular genomics studies. The role of skews and possibly of other sequence patterns in R-loop formation and the connection of the latter to the deposition (or its repression) of epigenetic marks are bound to be the focus of works to come.
Read more: A work published last year by the same group (Ginno et al. Molecular Cell 2012) introduces the concept of GC skews in the context of R-loop formation and their role in the maintenance of CpG island DNA composition.
 RSS Feed
RSS Feed
