point out a very important artifact of a very widely applied method, ChIPSeq. Both papers that appeared almost simultaneously show that there is an inherent bias in all ChIP experiments to result in enriching regions belonging to highly-expressed genes regardless of the nature of the studied protein.
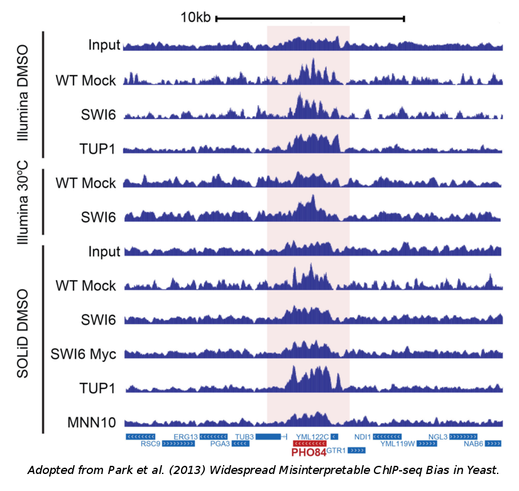
The data used: Both groups performed a series of experiments in yeast but their results may readily be extended to more complex genomes. Park et al. mostly focused on Tup1, a well-studied transcriptional supressor, the binding of which they surprisingly found to coincide with highly-expressed genes under multiple conditions. Teytelman et al. did the same starting from a family of repressors known as Sir proteins which they also saw to be consistently located in genes with high-expression.
The analysis: The analysis was pretty straight-forward. ChIPSeq data from various platforms were analyzed with standard peak-calling algorithms, such as MACS2 and the discovered peaks were correlated with the expression of the closest or overlapping genes. In all cases, a positive correlation was observed even if the protein under study was not a transcriptional activator. Teytelman et al. were able to show that a positive corrrelation was even observed in the case of a GFP ChIP experiment, that is a "mock" experiment with a protein that does not bind the DNA under normal conditions. The authors, thus, reach the conclusion that the major source of bias is the process of cross-linking which is likely to occur preferentially in actively transcribed regions.
What's next: Both papers point out a very important aspect that has largely escaped the attention of a huge number of published works. At the same time both groups suggest the performance of extensive controls for every ChIPSeq experiment to be performed in the future. For instance, performing a "mock" ChIP with a non-DNA-binding protein such as GFP and subtracting the signal (as representative of hyper-chipability) would distinguish signal from noise. This is bound to work to a great extent but is probably not cost-worthy, while it will complicate the downstream analysis through the addition of one more condition. On the other hand, new approaches that do away with cross-linking may provide more effective solutions.
Read more: A work published this week in Nature Methods by the group of S. Henikoff describing a methodology for inquiring transcription factor binding in native (non cross-linked) chromatin (Kazinathan et al. High-resolution mapping of transcription factor binding sites on native chromatin). This is probably the next step in this sort of approaches as native chromatin does away with cross-linking which is the main source of highly-expressed gene bias.
 RSS Feed
RSS Feed
