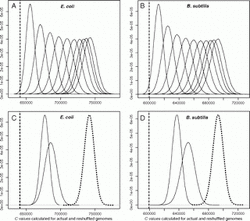
The data used: The complete genomes of E. coli and B. subtilis, their operon annotation (genomic coordinates and gene content) and the connections between operons and biological (mostly metabolic) pathways. Moreover, genes involved in pathways that are more active belong to operons that are located in smaller distances from each other.
The analysis: The authors devise a simple measure of operon compactness, that is how closely operons involved in the same pathway tend to occur in the linear genome. By comparing the actual average compactness value of E.coli and B. subtilis with one thousand random arrangements of operons (shuffling of the genome) they show that the natural arrangement is much more compact than the one that could have been produced by chance, therefore proposing that the observed compactness is the result of selection.
What's next: In a more recent paper (which we will be discussing soon), this idea is being further elaborated to incorporate data related to the 3-dimensional structure of the genome. It remains to be seen when and how such approaches will be extended to eukaryotic genomes with multiple chromosomes (and without operons)
Read more: Yin et al. (2010) Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome. PNAS
 RSS Feed
RSS Feed
